Billions of cells make up the fundamental building blocks of an organism, each performing its own functions. These cells work together like an incredibly complex, finely tuned integrated factory, maintaining the organism’s normal physiological functions. To conduct in vivo studies on different types and subtypes of cells, and to regulate the core functions of various cell types at the genetic level, it is essential to find highly targeted and efficient methods. Today, scientists use various biological techniques to deliver foreign genes into organisms via different vectors, such as recombinant adeno-associated virus (rAAV) vectors, to achieve precise, targeted regulation. This allows them to uncover the mechanisms behind various behaviors, physiological processes, and disease occurrences.
By mimicking the activation of endogenous genes—using promoters to drive the expression of exogenous genes—there is often a lack of targeting and delivery efficiency. Gene transcription and expression are regulated and influenced by multiple factors, and currently, we still cannot fully simulate the scenarios and conditions under which genes are driven to express in vivo. This results in promoter sequences being unable to truly achieve the efficiency and precision of endogenous gene expression. Against this backdrop, some binary-like regulatory system combination strategies have emerged. For example, scientists have introduced a site-specific recombinase (such as Cre and Flp) into target cells through gene knock-in methods. These recombinases can recognize specific DNA sequences and cleave those sequences. These sequences can be placed on either side of the exogenous gene we want to express. When we deliver the vectors into organisms to infect cells containing the recombinase, the recombinase can specifically recognize the sequences flanking the exogenous gene, cleave them, and cause the exogenous gene to flip or be deleted, thereby enabling the specific expression of the exogenous gene in target cells. Brain Case Biotech will explore the working principles of the recombinase systems in detail in the following sections.
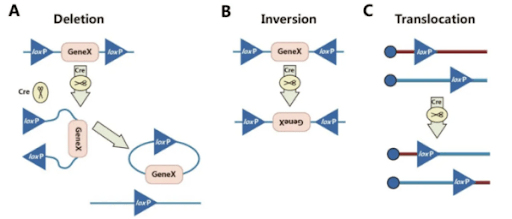
What are Cre and loxP?
In 1981, Nat Sternberg et al. reported a protein cloned from the P1 bacteriophage of Escherichia coli—a 38 kDa recombinase encoded by 343 amino acids called Cre (Cyclization Recombination Enzyme). It specifically recognizes loxP sites and mediates sequence deletion or recombination between loxP sites. The loxP (locus of X-overP1) site is 34 bp long, consisting of two 13 bp inverted repeat sequences and an 8 bp spacer region. The inverted repeat sequences are the specific recognition sites for the Cre recombinase, while the spacer region determines the orientation of loxP.
A. Deletion: When two loxP sites are on the same DNA strand and oriented in the same direction, the Cre recombinase excises the sequence between the loxP sites.
B. Inversion: When two loxP sites are on the same DNA strand and oriented in opposite directions, the Cre recombinase inverts the sequence between the loxP sites.
C. Translocation: When two loxP sites are on different DNA strands or chromosomes, the Cre recombinase induces an exchange between the DNA strands or a chromosomal translocation.
Classification of the Cre-loxP system
1. LSL Sequence (Cre-on)
Based on the above principles, the LSL sequence (loxP-STOP-loxP) was designed. In the absence of Cre recombinase, the downstream gene cannot be normally expressed due to the presence of the STOP cassette. Upon introduction of Cre recombinase, the STOP sequence is deleted, allowing the downstream gene to be expressed normally, thereby achieving Cre-dependent gene expression.
2. FLEX/DIO Sequence (Cre-on)
However, in practical applications, it was found that LSL could still lead to slight expression of the downstream gene even in the absence of Cre recombinase. In 2003, Frank Schnütgen et al. developed FLEX (Flip-excision) and, based on this, DIO (Double-Floxed Inverted Open reading frame), which changes the orientation of loxP and lox2272, greatly mitigating the leakage issue. lox2272 is also a commonly used lox site, similar to loxN, lox511, etc. lox2272 and loxP are mutually incompatible and are respectively targeted by Cre recombinase. In the DIO structure, Cre recombinase causes one inversion and one deletion through loxP and lox2272, thus achieving irreversible inversion and Cre-dependent expression of the target gene.
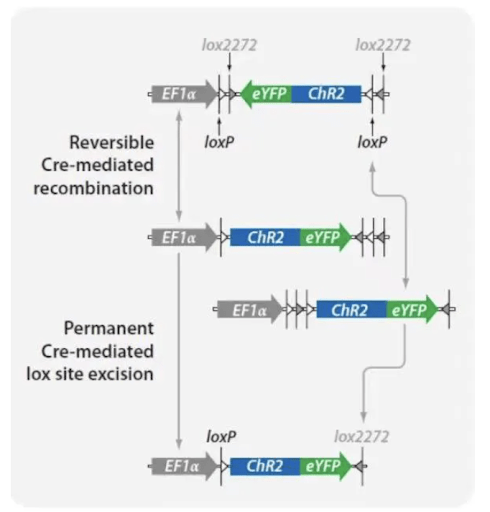
3. DO Sequence (Cre-off)
The inserted gene is oriented in the same direction as the promoter. In the presence of Cre recombinase, recombination occurs, causing the gene to invert and thus preventing normal expression.
4. CIAO Sequence (Cre-on)
Given the leakage expression issues with both LSL and DIO/FLEX, the Edward M. Callaway team designed a CIAO (cross-over insensitive ATG-Out) sequence, which almost eliminates the leakage problem. CIAO uses the ATG-OUT construction method, placing the ATG and the ORF of the target gene on opposite sides of the lox sequence. This means that if recombination does not occur, the target gene is incomplete, and even if transcription of the gene occurs, no functional protein will be translated. Additionally, the non-symmetrical lox66/71 sequences are used to further reduce spontaneous recombination during plasmid replication in bacteria. However, compared to loxP and lox2272, the recombination efficiency of Cre recombinase with lox66/71 is also reduced.

Application of the Cre-loxP system
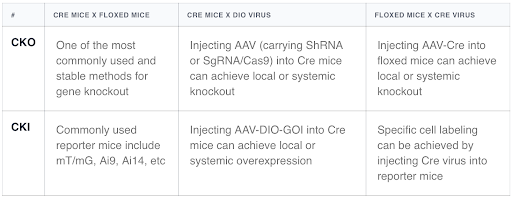
Why use viruses in addition to breeding two strains of mice?
This is because using viruses to achieve target gene overexpression (OE) or knockdown (KD) often results in a shorter experimental period and lower experimental costs compared to mouse breeding. Additionally, OE/KD resulting from mouse breeding is generally systemic, whereas viruses can more easily achieve gene regulation in local tissues or specific organs, allowing for more detailed studies.
Cross-usage of different recombinase systems
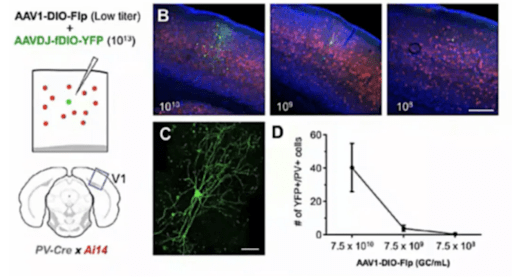
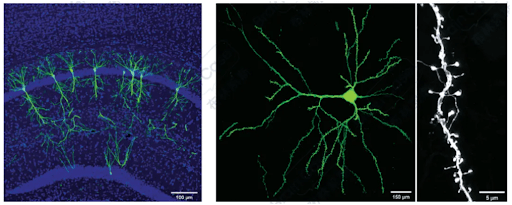
1. Sparse Labeling
By controlling the mixing ratio of AAV-DIO-Flp and AAV-FDIO-EYFP, the number of neurons can be controlled to achieve sparse and highly bright labeling. The team led by Xu Fuqiang at the Shenzhen Institute of Advanced Technology(Brain Case Biotech is the translational platform for the professor’s team, and all of these vectors were designed and provided by this platform).
They further optimized this method by adopting a cocktail packaging strategy, co-packaging pAAV-DIO-Flp and pAAV-FDIO-EYFP together, which reduced the issue of mutual exclusion during mixed co-infection of different batches of viruses.
With the continuous advancement of gene editing and viral vector technologies, scientists will be able to more precisely regulate cell functions and gain a deeper understanding of biological behaviors and disease mechanisms. This will pave the way for personalized medicine, precision therapies, and the development of new drugs.
In terms of technological applications, enhancing the efficiency and specificity of gene delivery while minimizing side effects will be a crucial focus of future research. As a key technological platform, Brain Case Biotech will continue to play a significant role in this field, providing more advanced tools and support for scientific research.